pgvector
Back when we wrote this, we were proud to bring HNSW indexes to Postgres. Since then, pgvector has added support for the HNSW index, and we've chosen to pause the development of pg_embedding and contribute to pgvector instead.
We plan on always supporting the latest version of pgvector. For more information, please refer to pgvector docs.
We’re excited to announce the release of our pg_embedding extension for Postgres and LangChain!
The new pg_embedding extension brings 20x the speed for 99% accuracy to graph-based approximate nearest neighbor search to your Postgres databases.
While the pgvector extension with IVFFlat indexing has been a popular choice, our new pg_embedding extension uses Hierarchical Navigable Small Worlds (HNSW) index to unlock new levels of efficiency in high-dimensional similarity search.
You can easily implement vector search with pg_embedding in your applications. Prior knowledge of vector indexes is optional. Run the following query to get started:
CREATE EXTENSION IF NOT EXISTS embedding;
CREATE INDEX ON items USING hnsw (embedding) WITH (maxelements = 1000000, dims=1536, m=32);You can also use the extension in LangChain using the PGEmbedding vectorstore:
from langchain.vectorstores import PGEmbedding
db = PGEmbedding.from_documents(
embedding=embeddings,
documents=docs,
collection_name="state_of_the_union",
connection_string=CONNECTION_STRING,
)
db.create_hnsw_index(max_elements= 10000, dims = 1536, m = 8, ef_construction = 16, ef_search = 16)For those curious about the inner workings and the differences between IVFFlat and HNSW for Postgres applications, we carried out benchmark tests on Neon Postgres to compare the performance of the two indexes. Keep on reading to find out more.
Benchmark Results
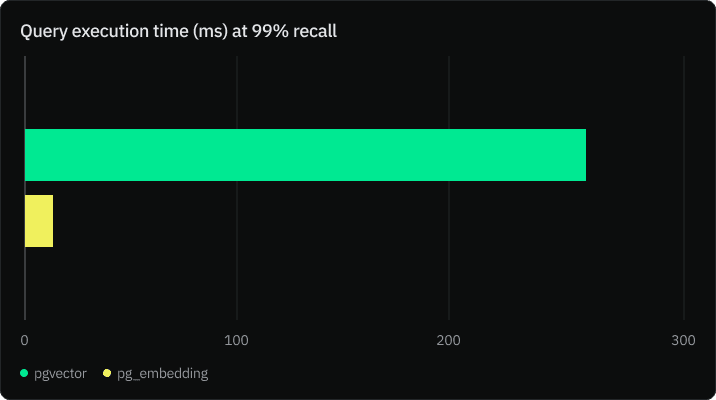
The benchmark tests compare the performance of pg_embedding with HNSW and pgvector with IVFFlat indexing using the GIST-960 Euclidean dataset, which provides a train set of 1 million vectors of 960 dimensions, and a test set of 1000. Each search returned k=100 vectors.
HNSW index is stored in memory for efficiency. For this to be a comparable test, we stored IVFFlat index in memory using pg_prewarm extension:
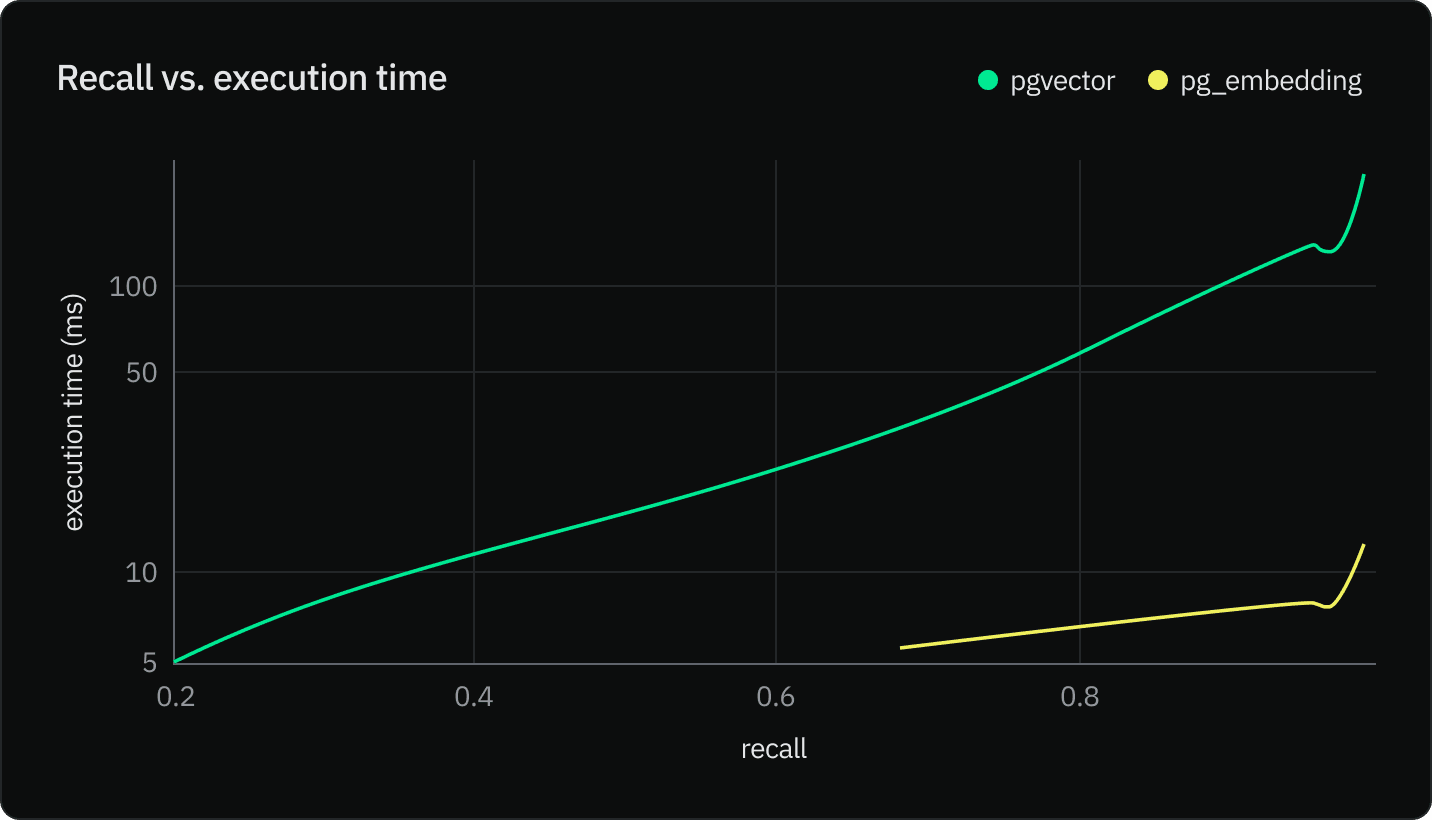
SELECT pg_prewarm('3448836', mode => 'buffer');The chart below is on a logarithmic scale. pg_embedding performs 5 to 30 times faster for the same recall. The higher accuracy we want to reach with IVFFlat, the longer the execution time. This is due the fact that more probes are required in pgvector and IVFFlat to reach larger recall. However, the bigger the number of probes, the faster search converges towards a sequential scan.
Results and Analysis
Metrics
We compared the performance of both extensions based on the following metrics:
- Execution time: Measured as the average time taken to perform 100 nearest neighbor search queries.
- Accuracy/Recall: Measured as the proportion of true nearest neighbors each query returns.
Setup
- Datasets: We used the GIST-960 Euclidean dataset for benchmarking. This dataset is widely used for benchmarking similarity search algorithms.
- Environment: The benchmarks were conducted on a Neon instance with 4 vCPUs and 16 GB RAM.
- Configurations:
- For pgvector, we varied the ‘probes’ parameter across [1, 2, 10, 50] and ‘lists’ parameter across [1000, 2000].
- For HNSW, we experimented with ‘m’ in [32, 64, 128], ‘efConstruction’ in [64, 128, 256], and ‘efSearch’ in [32, 64, 128, 256, 512].
Why is HNSW faster than IVFFlat?
Let’s start by understanding what IVFFlat and HNSW are and how they work.
IVFFlat with pgvector
pgvector allows for vector similarity search directly within the database. One of its indexing techniques is called IVFFlat. The IVFFlat index partitions the dataset into multiple clusters and maintains an inverted list for each cluster. During search, only a selected number of clusters are examined, which greatly speeds up the search process compared to a flat index.
By default, IVFFlat probes = 1, which means that the index will conduct the search in the nearest centroid’s cluster (or list). However, that can lead to inaccuracies as the query vector is closer to the cluster’s edges.
One way to increase accuracy is to increase the number of probes. The optimal number of lists and probes are respectively sqrt(rows), and sqrt(lists), which results in a time complexity of O(sqrt(rows)). The chart below shows that recall increases exponentially with the number of probes.
HNSW and pg_embedding
HNSW (Hierarchical Navigable Small World) was first introduced by Yu A Malkov and Dmitry A Yashunin in their paper titled: Efficient and Robust Approximate Nearest Neighbor Search Using Hierarchical Navigable Small World Graphs.
HNSW is a graph-based approach to indexing high-dimensional data. It constructs a hierarchy of graphs, where each layer is a subset of the previous one, which results in a time complexity of O(log(rows)). During search, it navigates through these graphs to quickly find the nearest neighbors.
There are three main parameters in HNSW algorithms:
- m: This parameter refers to the maximum number of bidirectional links created for every new element during the construction of the graph.
- efConstruction: This parameter is used during the index building phase. Higher efConstruction values lead to a higher quality of the graph and, consequently, more accurate search results. However, it also means the index building process will take more time. The chart below shows build time by ef_construction value using the GIST-960 dataset.
- efSearch: This parameter is used during the search phase. Like efConstruction, a larger efSearch value results in more accurate search results at the cost of increased search time. This value should be equal or larger than k (the number of nearest neighbors you want to return).
By default, efConstruction and efSearch are 32, but you can modify their values when you create the HNSW index:
CREATE INDEX ON vectors_hnsw USING hnsw (vec) WITH (maxelements = 1000000, dims=1536, m=32, efconstruction=32, efsearch=32);Which index should you pick?
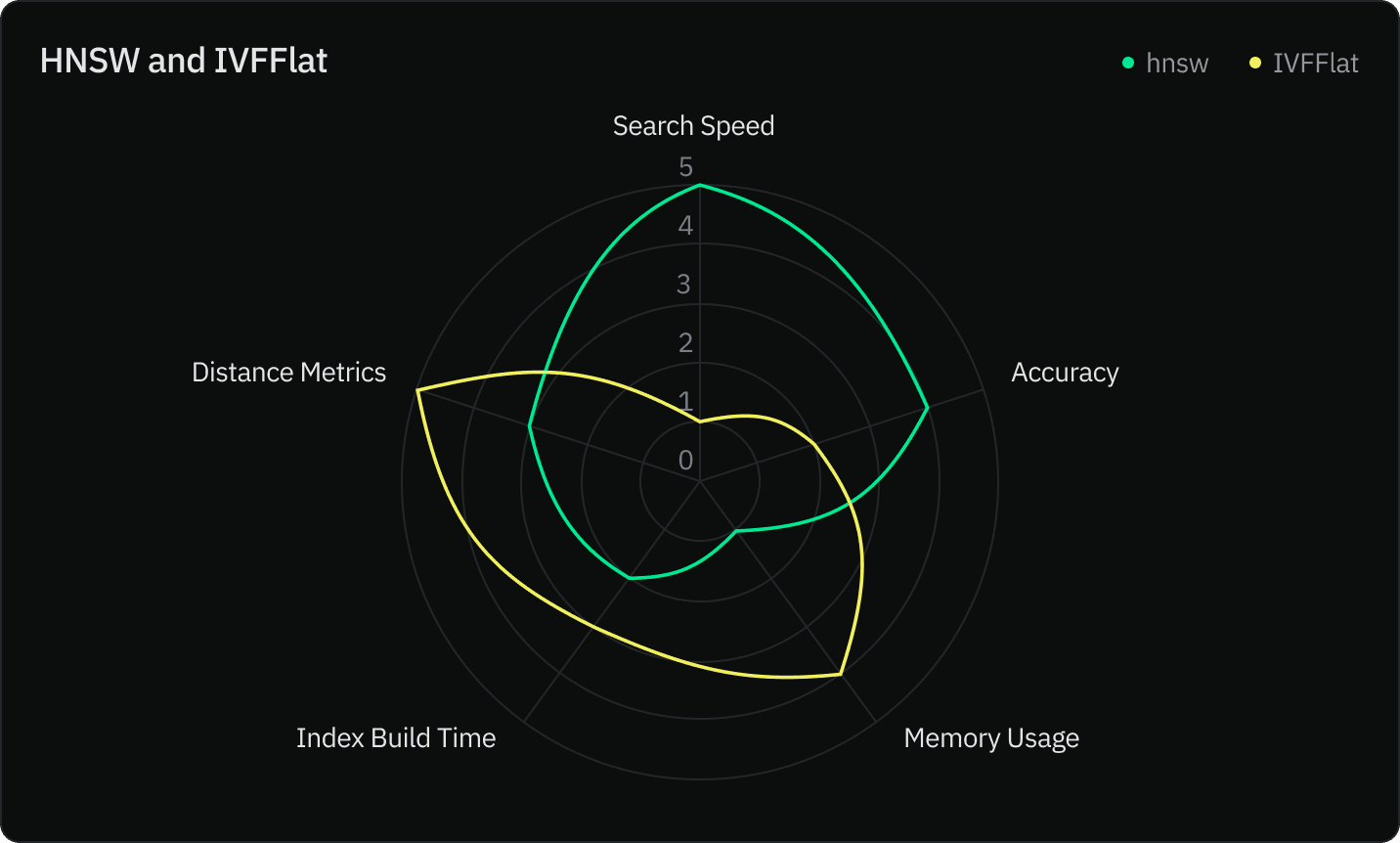
We compared both indexes using five criteria:
- Search speed
- Accuracy
- Memory usage
- Index construction speed
- Distance metrics
| IVFFlat | HNSW | |
| Search Speed | Fast, but the search speed depends on the number of clusters examined. More clusters mean higher accuracy but slower search times. | Typically faster than IVFFlat, especially in high-dimensional spaces, thanks to its graph-based nature. |
| Accuracy | Can achieve high accuracy but at the cost of examining more clusters and hence longer search times. | Generally achieves higher accuracy for the same memory footprint compared to IVFFlat. |
| Memory Usage | It uses relatively less memory since it only stores the centroids of clusters and the lists of vectors within these clusters. | Generally uses more memory because it maintains a graph structure with multiple layers. |
| Index Construction Speed | Index building process is relatively fast. The data points are assigned to the nearest centroid, and inverted lists are constructed. | Index construction involves building multiple layers of graphs, which can be computationally intensive, especially if you choose high values for the parameter ef_construction |
| Distance Metrics | Typically used for L2 distances, but pgvector supports inner product and cosine distance as well. | Only uses L2 distance metrics at the moment. |
Your choice between the `pg_embedding` and the pgvector depends on your specific use case and requirements:
- Memory Constraints (
pgvector): If you are working under strict memory constraints, you may opt for the IVFFlat index as it typically consumes less memory than HNSW. However, be mindful that this might come at the cost of search speed and accuracy.
- Search Speed (
pg_embedding): If your primary concern is the speed at which you can retrieve nearest neighbors, especially in high-dimensional spaces, pg_embedding is likely the better choice due to its graph-based approach.
- Accuracy and Recall (
pg_embedding): If achieving high accuracy and recall is critical for your application, pg_embedding may be the better option. HNSW’s graph-based approach generally yields higher recall levels compared to IVFFlat.
- Distance Metrics (
pgvector): Bothpgvectorandpg_embeddingsupport L2 distance metric. Additionally,pgvectorsupports the inner product, and cosine distance.
Conclusion
With the introduction of the pg_embedding extension for Postgres, you now have a powerful new tool at your disposal for handling high-dimensional vector similarity searches efficiently within your database. The graph-based nature of the HNSW algorithm offers several advantages over the IVFFlat index in terms of search speed, accuracy, and ease of setup.
IVFFlat index with pgvector remains a viable choice for applications with stringent memory constraints but at the expense of recall.
Ultimately, the choice between pg_embedding and pgvector with IVFFlat should be informed by the specific demands of your application. We encourage you to experiment with both approaches to find the one that best meets your needs.
We are excited to see the innovative applications you will develop with pg_embedding and look forward to your feedback! Stay tuned for further updates and enhancements.
📚 Continue reading
- Understanding vector search and HNSW index with pgvector: learn how HNSW enhances the efficiency and responsiveness of AI applications by allowing faster and more scalable vector similarity searches.
- What you need to know about Mixtral 8x7B: learn about the advancements introduced by this model, including potential cost benefits.
- 30x faster index build for your vector embeddings with pgvector: learn how the new pgvector speeds up the index building process for vector embeddings by 30 times, optimizing performance for your AI apps.